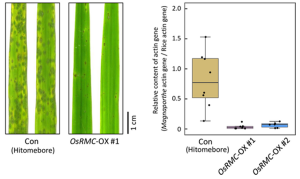
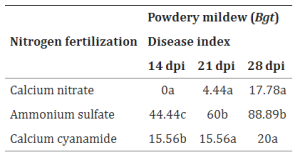
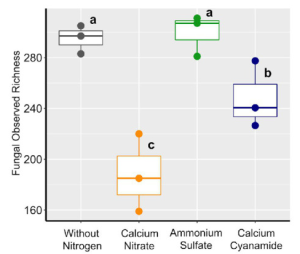
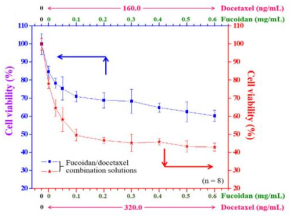
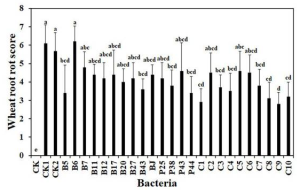
Continuous cropping of Sugar beet changed rhizospheric fungi significantly than non-cropping
A group from National Sugar Crop Improvement Centre, Heilongjiang University, Harbin, China, etc. has reported about difference of rhizosphere between continuous and non-continuous cropping groups of sugar beet rhizosphere.
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC9490479/
There were significant differences in fungal community composition between continuous and non-continuous cropping groups of sugar beet rhizosphere.
Compared with non-continuous cropping, continuous cropping increased the relative abundance of potentially pathogenic fungi such as Tausonia, Gilbellulopsis, and Fusarium, but decreased the relative abundance of Olpidium.
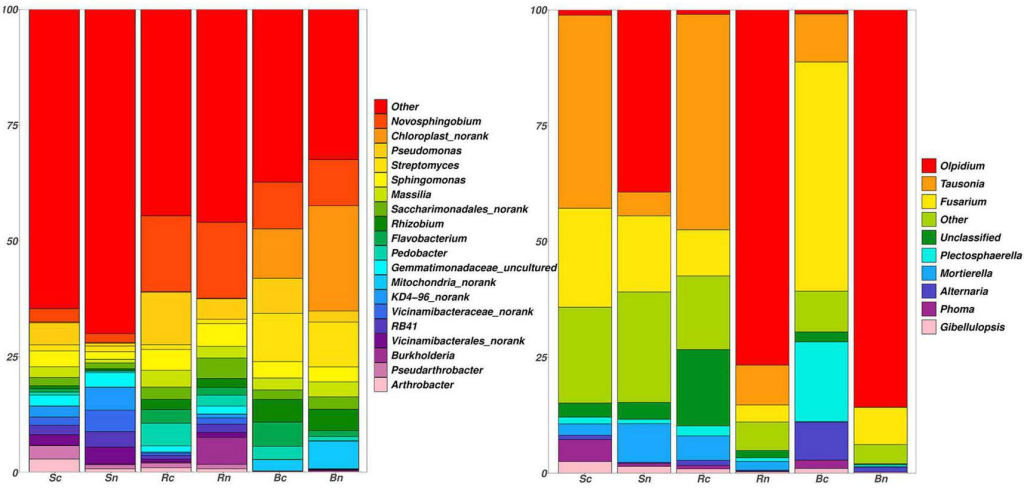
Left figure=rhizospheric bacteria, Right figure=rhizospheric fungi
where, Sc, continuous cropping bulk soil; Sn, non-continuous cropping bulk soil; Rc, continuous cropping rhizosphere soil; Rn, non-continuous cropping rhizosphere soil; Bc, continuous cropping sugar beetroot; Bn, non-continuous cropping sugar beetroot.