A lectin domain exists in mouse N-acetylglucosaminyltransferase-IVa (MGAT4A: GnT-IVa) C-terminal region
A group from Department of Molecular Immunology, Research Institute for Microbial Diseases, Osaka University, Japan, etc. has discovered a lectin domain in mouse N-acetylglucosaminyltransferase-IVa (MGAT4A: GnT-IVa) C-terminal region.
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC9296478/
The biosynthesis of the GlcNAc branches existing in N-glycans is catalyzed by the specific N-acetylglucosaminyltransferases (GnTs), GnT-I to -V.
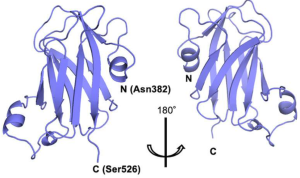
Authors have found that the lectin domain discovered in GnT-IVa forms a β-sandwich fold composed of nine β-strands with three short α-helices, and this domain shows structural and functional similarities with bacterial CBM32 acting as a lectin with a preference for GlcNAc.
It was also found that the lectin domain of GnT-IVa is required for the efficient N-glycan biosynthesis toward glycoprotein substrates in cells.