Interactions between galectins and O-mannosylated core M1 glycopeptides of α-dystroglycan
A group from Frontier Research Center for Advanced Material and Life Science, Hokkaido University, Sapporo, Japan, etc. has reported about interactions between galectins and O-mannosylated glycopeptides of α-dystroglycan, especially focusing on its core M1 structure.
https://www.nature.com/articles/s41598-022-22758-0
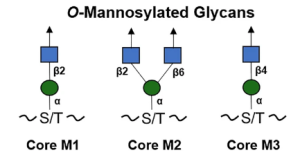
The O-linked mannose (O-Man) exists in a limited number of proteins that are required for normal development and have vital functions in muscle and neural physiology. The α-dystroglycan (α-DG) is the extracellular component of dystroglycan (DG), and is the most extensively studied mammalian O-Man glycoprotein. It is ubiquitously expressed in the skeletal muscles and the brain and is associated with cell adhesion, muscle integrity, and neurological development. α-DG possesses unique glycans, LacNac-terminated three kind of core structures (M1, M2, and M3), in its mucin (MUC)–like domain.
In this study, it was shown that Human Gal-1, -4, and -9 (except -3) can strongly recognize O-Man LacNAc-terminated glycoconjugates, and the presence of an α2,3-sialylated terminus led to a major reduction in the affinity of galectin, suggesting that this type of extension can fine-tune galectin activity towards this type of O-Man glycans. These interactions were significantly inhibited by lactose, establishing that the α-DG core M1-type glycans bind to the canonical sugar-binding site (S-face) of galectin, thus serving as a receptor for galectins.
And further, it was shown in microarray experiments that Gal-1 revealed trans-bridging capabilities, linking laminin-111, -121, -211, and -221 (but little -511) and core M1 α-DG glycopeptides as shown below, providing a new insight on the therapeutic application of this galectin in muscular dystrophy.
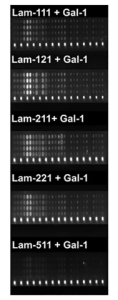
Fluorescence images of M1 glycoconjugates microarrays with laminins plus galectins