
- Glycan Profiler (GlycoStation)
- Glycan Profiling Analysis Softwar (ToolsPro)
- Statistical Analysis and Deep Learning Softwar (SA/DL-Easy)
- Culture Media (Domestic Service only)
- Lectin Microarray (LecChip)
- Standard Protocol for LecChip
- Lectin (SeviL, MytiLec etc.)
- Optic Biome Senso (OBS)
Glycan Profiler and Glycan Profiling Analysis Software
Glycan Profiling using Lectin microarrays

4th Anniversary: Half price sale on GSR2300, special sale for 1 unit only
Glycan Profiler is a kind of microarray scanner. This naming was given to emphasize that it is possible to get high fidelity Glycan Profiling. And, GlycoStation® is the name of our platform for Glycan Profiling Analysis. GlycoStation® Reader is a scanner in this platform, and was designed to be able to take Glycan Profiles of glycoproteins by using Lectin microarrays. GlycoStation® Reader is possible to detect very weak glycan-lectin biomolecular interactions non-destructively with using an evanescent-field fluorescence excitation technology. Accordingly, it is very strong in Glycan Profiling Analysis, but is able to scan Glycan array nondestructively as well. GlycoStation is the registered trademark of Mx.
Reference) Evanescent-field fluorescence-assisted lectin microarray: a new strategy for glycan profiling, Atsushi Kuno, Noboru Uchiyama, Shiori Koseki-Kuno, Youji Ebe, Seigo Takashima, Masao Yamada & Jun Hirabayashi, Nature Methods (2005)
Development of an ultra-high sensitive and high-speed glycomic profiling scanner (Joint Development with AIST)
Prominent features of GSR2300
The current high-end Glycan Profiler is GlycoStation® Reader 2300 (GSR2300). And, GlycoStation® Reader 2200 (GSR2200) was a sibling of the GSR2300 and has been integrated with the GSR2300 now. Comparing GSR2300 with the first generation Glycan profiler GSR1200, the prominent feature is that two contradictory functions (i.e., high sensitivity and fast scanning) are satisfied simultaneously in GSR2300.
It is able to scan microarray surface in only 15 seconds with ultra-high sensitivity. Taking into consideration that the common microarray scanners need around 10 minutes for the scanning, GSR2300 would be a world fastest scanner with ultra-high sensitivity. Utilizing its low noise detection capability, the linearity in a lower signal range is also greatly improved, and thereby the dynamic range in lower signal side is greatly expanded.
Faster is always better. Please have an experience with the world fastest and the ultra-high sensitivity scanner GSR2300. You will be unable to get back to other scanners, if you experience such high speed scanning with this.
Featuring Specs of GSR2300

- Evanescent-field fluorescence excitation technology: High sensitivity detection of weak biomolecular interactions with a non-destructive way
- 3 colors with Cy3, Cy5, FITC as a standard: 8 kinds of optical filters can be installed at the maximum
- Ultra high sensitivity: Glycan profiling from only 3 cells
- Low noise: S/N is improved by 60 times at the maximum comparing with GSR1200 (depends on samples)
- Digital Binning function: 2×2 mode, 4×4 mode
- Super high speed scanning: 15 seconds (at the maximum sensitivity with Digital binning)
- Scan Area: Standard 12mm x 65mm, and Max 24mm x 65mm. The default scan area is 12mm x 65mm to match the LecChip format. The scan area can be changed to the maximum of 24mm x 65mm. This change is possible just by rewriting the GSR operation definition file. No particular mechanical changes to the device are required.
- Resolution: 6.5um
- Picture formats: TIFF, JEG, BMP
- Size: 440x592x585mm
- Power supply: 100V ~ 240V
- Weight: 67kg
- Periodic Replacement Part:Metal Halide Bulb (every 1,500 hours)
- Sales Price: Please contact us (info@emukk.com)
Scanned images: High resolution and Low distortion
In order to satisfy above listed specs, the optical system was redesigned completely, and a proprietary lens (named GT lens) with high NA and wider field size is used in GSR2300. The detailed explanation about the performance of GSR2300 is written in the following link and a latest paper as a reference.
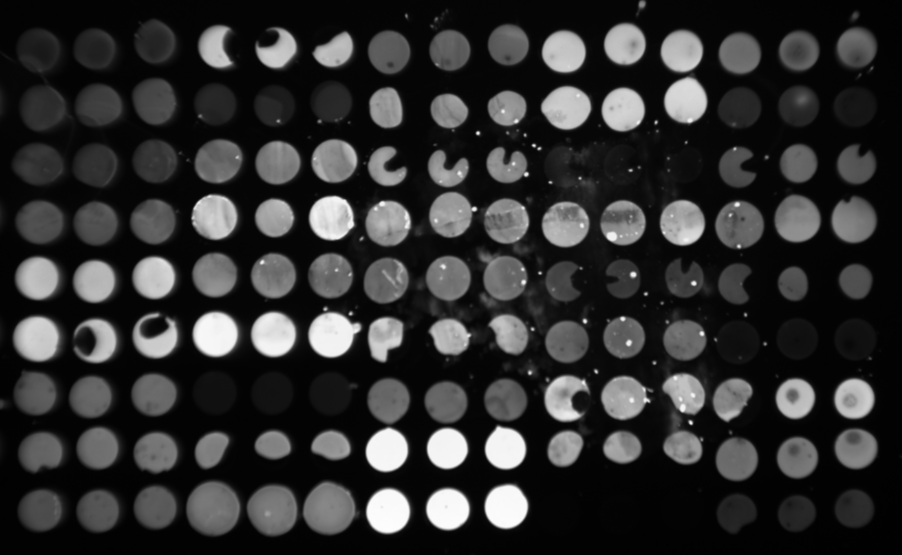
Detailed explanation about the performance of GSR2300
Reference)A latest paper about the high sensitivity of GSR2300, An improved evanescent fluorescence scanner suitable for high-resolution glycome mapping of formalin-fixed paraffin-embedded tissue sections, Patcharaporn Boottanun, Chiaki Nagai-Okatani, Misugi Nagai, Umbhorn Ungkulpasvich, Shinjiro Yamane, Masao Yamada & Atsushi Kuno, Analytical and Bioanalytical Chemistry (2023) . In this paper, GSR2300 was called as mGSR1200-CMOS scanner.
Ultra-high speed, High sensitive Glycan Profiler
We are pleased to inform you of the release of GlycoSuperLiteTM 2200 (GSL2200: US Model # GSL1150), the next generation of GlycoLite. The former GlycoLite 2200 (GL2200) has been already discontinued. GlycoSuperLiteTM must be the long-awaited basic glycan profiler with greatly improved appearance and performance. This device is perfect for quick analysis, where you want to know the changes in glycan modification with comparative glycan profiling rather than detailed glycan analysis using LC/MS-MS.

This unit uses a reduction optical system by 5.5 times with high NA, and can instantly capture fluorescent images of lectin microarrays without scanning. Thereby, ultra-high-speed scanning of 10seconds or less is attained. It is no mistake to say that it is the world’s fastest glycan profiler adopted the evanescent-field fluorescence excitation technology. When you actually use it, you will be amazed at its high speed, which does not apply to the concept of a scanner. But, the sensitivity is not sacrificed even in this high-speed scan, and the detection limit sensitivity reaches 2 ng/mL.
Due to the adoption of the reduction optical system, the resolution is reduced to some extent. However, in microarrays with a pitch of around 500 ~ 600 um, spot identification and analysis can be performed without problems. The miniaturization of the device has also progressed, and the weight of the main body is 10 kg, which is less than half the weight of the former GlycoLite.
- Evanescent-field fluorescence excitation technology: High sensitivity detection of weak biomolecular interactions with a non-destructive way
- Fluorescence: Cy3
- High sensitivity: LOD=2 ng/mL
- Super high speed scanning: 10 seconds (at MAX sensitivity), 133msec (Fastest)
- Picture formats: TIFF, JPEG, BMP
- Scan Area: Standard 25mm x 75mm
- Power Supply: 100V ~ 240V
- Size: 350x280x350mm
- Weight: 10.4kg
- Sales Price: Please contact us (info@emukk.com)

It’s cheaper if you buy this as your own tool, comparing with contract analysis service
The list price of GSL2200 is to be announced, but low price that does not disappoint you definitely. Comparing purchasing GSL2200 with using the contract analysis service, the former case will be cheaper than the latter case, when the number of analysis samples exceeds 30.
Spec. comparison among GSR2300, GSL1150, and discontinued GL2200
Significance of GSR2300/GSL2200
Commercially available microarray scanners generally have a resolution of 4 um to 100 um. Most existing scanners are laser scanning type, and the scanning time is around 5 to 10 minutes with a typical resolution of 10 um. In the case of laser scanning type scanners, the resolution and the scanning time are inversely proportional, so the higher the resolution, the longer the scanning time is, up to 20 min. and further 30 min. or more.
The resolution of GSR is 6.5 um, and the scan time is Max 80 seconds and Min 10 seconds under standard 1×1 mode. On the other hand, GSL has a resolution of 35um, and a scan time is Max10 seconds and Min 133 msec.
The resolution of these models is constant.
The biggest difference between general scanners and GSR/GSL is the excitation method. The use of evanescent-field fluorescence excitation technology is the biggest differentiating factor.
The benefits of GSR/GSL can be summarized as follows.
- using evanescent-field fluorescence excitation technology, weak biomolecular interactions can be measured non-destructively and with high precision,
- high sensitivity and high-speed scanning coexists,
- if the spot pitch is around 500 ~ 600 um, there are practically no problems even at GSL resolution.
- the fact is that once people get used to speed, they can’t go back.
Resolution of GSL
The resolution of GSL is lower than that of GSR due to the adoption of a reduction optical system, but in practice, the spots are clearly separated as shown in the figure below, and we would like to say that there is no problem in degitizing those spots with ToolsPro. The array used for the image acquisition has abnormal spot shapes as shown below, but we ask for your understanding in using it just as a test chip for the image acquisition testing.

Glycan Profiling Analysis Software
Glycan profiling analysis software, GlycoStation® ToolsPro, is attached in GSR2300 as a standard. The current version is 3.0 (abbreviated name, ToolsPro Ver3.0). It is designed as a user friendly software equipped with all of necessary functions in performing glycan profiling analysis. The featuring specs are as follows.
- New microarray formats are installed in addition to the current standard microarray format 45lectins/7wells (90lectis/3wells and 18lectins/14wells)
- Generate grids on scanned images (with TIF format) and digitize spot intensities (16bits)
- Selection of two background modes (Local Background mode and Global Background mode)
- Expand dynamic range in a higher signal range using a gain integration (data integration) function with two or more fluorescent images taken at different gains (exposure times), and thereby compensating saturated lectin spot intensities
- Draw binding curves (sample concentration on transverse axis, and net intensity on longitudinal axis), easy to select the most appropriate sample concentration
- Draw comparative glycan profiling pictures (Max 10 comparisons)
- Data normalization (Raw, Max Intensity, Average, Special Lectin)
- Copy and paste drawings onto other software such Powerpoint through clipboard
- Export numerical data used for drawings onto Excel
- Export data sets onto statistical software and deep learning software (GlycoStation® SA/DL Easy)
- Edit lectin names, in order to deal with any lectin panels
This software is protected by a dongle key.

Download Link of old version Sofware
GlycoStation ToolsPro Ver1.5-Ver2.0 Installer
Sentinel System Driver 7.4.2 Installer
Software
Statistical analysis & Deep Learning Software (GlycoStation® SA/DL Easy)
Any software knowledge does not be required in using SA/DL Easy. With this software, a neural network is easily created by just clicking mouse and selecting items, and you could experience the world of Deep Learning so easily. The data input format is a csv file. Please enjoy the AI world with this user friendly software.
Let us introduce you a good example of this application. A group from the National Institute of Child Health and Development demonstrated that deep learning can be combined with glycan profiling data from lectin microarrays to recognize cell differences with ultra-high accuracy.
Recognition of cell types with ultra-high accuracy and non-destructive way
The lectin microarray used was LecChip Ver1.0 of GlycoTechnica (which was developed and marketed by Mortex Glycomics Laboratory). The lectin microarray has 45 kinds of carefully selected natural lectins and has been widely used worldwide as a de facto standard for lectin arrays since its launch in 2007.
Deep Learning, as you know, is now used in various fields as one of methods of AI, and in this research, Google’s TensorFlow was used as the backend, and Keras was used as wrapper software. The layer configuration of this deep Leaning had an input layer of 45 (the same number of lectins) and an output layer of 5 (to discriminate five cells), and hidden layer from 1 to 5. There were five types of cells evaluated (Pluripotent stem cell, Mesenchymal stromal cell, Endometrial and ovarian cancer cell, Cervical cancer cell, Endometrial cell), and a total of 1,577 samples were used for the evaluation. The results were astonishing, and showed a high recognition accuracy of 97.4% overall
Consumables: R&D use
Culture Media
Culture media are now for domestic service only. That is because it must be kept in cold storage including the transportation periods from here to overseas countries. Under such conditions, shipping cost from here to overseas countries is not insignificant, comparing to the price of culture media itself. We are very sorry for the inconvenience and appreciate your understanding.

A new product released, March 6th, 2021: EMUKK-35 (Special culture medium) especially for fibroblast.
Three new products released, May 15th, 2021: EMUKK-05 (Special culture medium) especially for hepatocytes,
EMUKK-15 (Special culture medium) especially for epithelial cells,
EMUKK-25 (Special culture medium) especially for mesenchymal stromal cells.
Detailed information about Hepatocytes and its recommended culture media is as follows.
Hepatocytes (HepaMN cells, ProliHH cells) and its recommended culture media
Lectin Microarray
There are two types of lectin microarrays, LecChip Ver1.0 developed and marketed by Moritex Glycomics Laboratory and LecChip Ver2.0 (which adopted the same format as LecChip Ver1.0) developed and marketed by GlycoTechnica. The compatible products will be manufactured and supplied by our partner company, Precision System Science Co., Ltd.. Please contact us if you have any questions about it.
The lectins immobilized on Ver1.0 and Ver2.0 and their glycan binding specificities are as follows (pdf).
Ver1.0 has followed the same lectins since Moritex’s glycomics laboratory launched it in 2007, and it is highly comprehensive, so it is no exaggeration to say that it is the world’s standard lectin microarray. On the other hand, Ver2.0 adopts recombinant lectins greatly, and the new lectins which recognize high mannose-type glycan structures (Type I to IV), Core fucose, and α-Gal are enriched. Together with Ver1.0 and Ver2.0, glycan profiling using 83 kinds of lectins is possible. LecChip was the registered trademark of GlycoTechnica., and then it was inherited by PSS.

LecChip Ver1.0-Specificity.pdf

LecChip Ver2.0-Specificity.pdf

Lectin
Special lectins will be sales released as our new products.

The first one will be SeviL which has binding specificity to GM1b and GA1, and the second one is MytiLec which has binding specificity to disaccharide melibiose, Galα(1,6)Glc and the trisaccharide globotriose, Galα(1,4)Galβ(1,4)Glc.
Sucessively, HOL-30 has binding specificity to both Type-1 (β1-3) and Type-2 (β1-4) LacNAc. There is no known lectin which has affinity to Type-1 LacNAc, and existing lectins such as ECA, RCA120, PHAE, PHAL bind only to Type-2 LacNAc. By using HOL30, it is possible to detect Type-1 LacNAc in addition to Type-2 LacNAc.
The next coming hRTL has binding specificity to mucin-type glycan TF-antigen. There are some existing lectins which bind to TF-antigen, that is ABA, ACA, and PNA.
| Lectin names | Glycan binding specificities |
| SeviL | GM1b, GA1 |
| MytiLec | Galα(1,6)Glc, Galα(1,4)Galβ(1,4)Glc |
| HOL-30 | Type-1 and Type-2 LacNAc |
| hRTL (Coming Shortly) | TF-antigen |
Glycan binding specificity of SeviL and its molecular structure analysis
Glycan binding specificity of MytiLec and its molecular structure analysis
Computationally designed Mitsuba lectin based on MytiLec-1
Glycan binding specificity of HOL-30



New Biosensors

Biosensing, Bioimaging Insturuments
Vaccination for the new coronavirus (COVID-19) is gradually progressing in the world. I think it will take some time, but the pandemic will be over with prevailing vaccination in a couple of years. With advent of various SARS-CoV-2 variants, the efficacy of current vaccines would be reduced. However, it does not always mean that the current vaccines do not work at all, and probably a boost vaccine against such variants will come out.
Gene amplification is essential to get enough detection sensitivity, and therefore, RT-PCR continues to exist as the gold standard. However, in order to speed up and simplify genetic testing further, other innovative methods using isothermal gene amplification combined with CRISPRA-Cas12a, -13a have been developed.
From a view point of POCT, however, small and inexpensive tests are required. Various devices are coming out as prototypes, but the protocol tuning is still very important in terms of speeding up testing. As a detection method itself, an electrical detection method would be probably the most flexible. From that point of view, I think that an electrochemical detection method using the Redox reaction would be the most interesting.
Comprehensive Review on Elecrochemical detection of Glycoproteins
Optic Biome Sensor (OBS)
We are planning to develop a novel rhizospheric optic biome sensor. The fundamental patent for the optic biome sensor (OBS) has been already applied on Sept. 14th, 2022 in Japan (2022-146003). You could see some results for measuring Bacillus and Pseudomonas bacteria.

One of the good application examples of OBS will be rhizosperic microbiota. Rhizobacteria coexist around the roots of plants, and the diverse and complex population of bacteria is called the rhizospheric microbiota. This symbiotic relationship between roots and rhizobacteria is often compared to the relationship between animal intestines and gut microbiota.
In addition to the traditional indicators used in soil diagnosis, chemical indicators such as pH and water content, and physical indicators such as soil density, there is an increasing need for sensors that can detect biological features.
Recent advances in gene analysis technology have made it possible to analyze the composition of the rhizospheric microbiota without the need for isolation and culture of rhizobacteria from soil. The interaction between individual bacteria is also being understood, for instance, in the form of helper bacteria that help mycorrhizal bacteria, and rhizobacteria fix nitrogen and solubilize nutrients (phosphorus, potassium, etc.) in the soil. In addition to exhibiting such functions, it secretes antibacterial substances against phytopathogens and phytoholmons into the rhizosphere as metabolites, greatly contributing to soil fertility.
From this point of view, there is a need for a rhizospheric optic biome sensor that captures an accurate indicator of the health of the rhizosphere of the plant, and it would be the best to find a way to control soil fertility using it.
Technical Support and Consulting
Utilizing Mx’s wide range of experience, Mx is able to support a variety of products development such as microarray scanner, microarray, biomarker discovery, and its diagnostic assay.
