Rhizosphere of red maize: Effective pathogen inhibitory substances were discovered from Pseudomonas protegens EMM-1
A group from Benemérita Universidad Autónoma de Puebla (BUAP), Puebla, Pue., México, etc. has reported on pathogen inhibitory substances extracted rhizospheric bacteria, Pseudomonas protegens EMM-1.
https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0240545
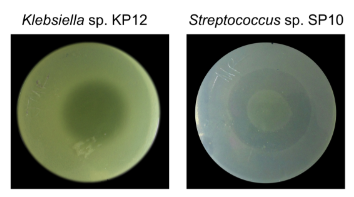
Pseudomonas protegens EMM-1 was isolated from the rhizosphere of red maize. It was found that P. protegens EMM-1 can efficiently inhibit the growth of a broad spectrum of bacteria, including Klebsiella sp. and Streptococcus sp.
To evaluate the inhibitory effect of secreted substances, First of all, conventional centrifugation methods for obtaining cell-free culture supernatants (CFSs) were used. However, CFSs from P. protegens EMM-1 did not show inhibitory activity. Instead of that, the inhibitory substances could be isolated from agar by the cold-leaching methodology using ethanol. The inhibitory substances displayed strong inhibitory activity as shown below, which suggests the existence of other inhibitory substances that could be explored in the future.
The molecular size of the inhibitory substances were estimated between 3 to 10 kDa. The inhibitory activity was completely lost at 100°C but remained stable between -4°C and 60°C. The inhibitory activity of the crude extract was stable within the pH range of 6 to 8. In this paper, the molecular structures of those inhibitory substances were not identified unfortunately.
Anyway, this work demonstrated the significance of evaluating rhizospheric bacteria to discover inhibitory compounds that may potentially be used for the development of new antimicrobial therapies for application in medicine or agriculture.