Glutamic acid directly modulates the composition of the microbiome community of strawberry and tomato
A group from Gyeongsang National University, Jinju, Korea, etc. has reported that glutamic acid directly modulates the composition of the microbiome community of strawberry and tomato.
https://pubmed.ncbi.nlm.nih.gov/34930485/
Throughout plant development, the structure of the associated microbial community has a significant impact on plant health, especially in disease prevention by beneficial microbes such as Bacillus, Pseudomonas, and Streptomyces.
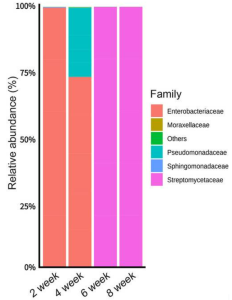
Whether l-glutamic acid can modulate the structure of the strawberry anthosphere microbial community was examined. Each of three treatments (untreated control, l-glutamic acid, or l-asparagine at a final concentration of 2%, pH 6.5) was sprayed for 1 min per plot at 2-week intervals. Enterobacteriaceae dominant from weeks 2–8 in the untreated control, in the l-asparagine treatment, and from weeks 2–4 in the l-glutamic acid treatment. Pseudomonadaceae were the second most abundant species, followed by Moraxellaceae. Enrichment of Streptomycetaceae occurred only in the l-glutamic acid treatment during weeks 6 and 8, accounting for 99.9 and 99.9% of the community (see below).
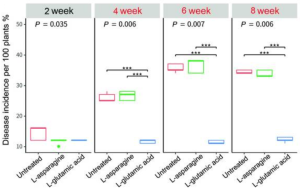
Then, the effect of l-glutamic acid on the occurrence of gray mold, blossom blight, and the density of Streptomyces globisporus SP6C4, in strawberry flowers was studied. Disease incidence (DI) was evaluated at 2-week intervals. At the same time, l-glutamic acid and l-asparagine were sprayed three times, at 2-week intervals, from week 4 to week 8. Gray mold DI from week 0 to week 4 remained relatively low (10–16%) regardless of treatment. At 6 weeks, the untreated control presented a DI of 16.6%; the DI in the l-asparagine-treated plot was 16%, and the DI in the plot treated with l-glutamic acid was significantly lower, at 12.4%. At week 8, the DI in the untreated control increased to 34% but that in the plot sprayed with l-glutamic acid was maintained below 17% (see below).
Thus, it was shown that glutamic acid reshapes the plant microbial community and enriches populations of Streptomyces, a functional core microbe in the strawberry anthosphere. Similarly, in the tomato rhizosphere with more complex microbiome, treatment with glutamic acid increased the population sizes of Streptomyces as well as those of Bacillaceae and Burkholderiaceae. At the same time, diseases caused by species of Botrytis and Fusarium were significantly reduced.