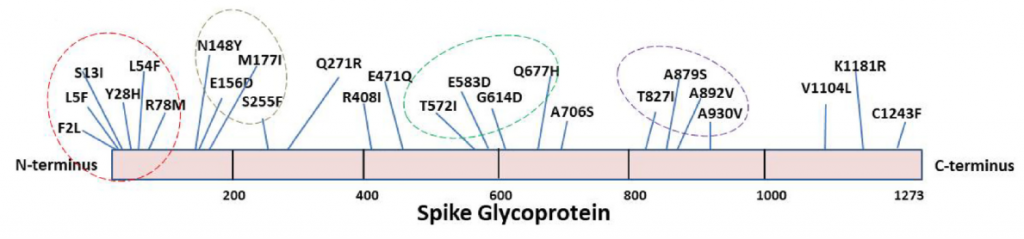
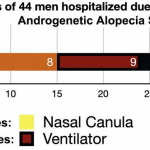
Mutations in S-proteins of the new coronavirus (SARS-CoV-2): 25 mutations, including A930V, D614G, A706S, and A879S
From the RNA sequence of the new coronavirus (SARS-CoV-2) registered from India in NCBI-Virus-database as of June 6, 2020, a research result has been reported on what mutations occurred compared to the original strain that began to spread in Wuhan, China.
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7521409/
There are 25 mutations, and they are divided into four clusters (1-100, 148-255, 570-680, 820-930).
Mutations in T572I, A879S, A892V, and A930V have caused significant changes in the secondary structure of proteins, T572I causes a structural change to coilded→helix and the other three to helix→beta sheet. Mutations in the A930V, D614G, A706S, and A879S seem to make a relatively large difference in protein structure. Of particular interest is that the mutation of D614G occurs in 88% of RNA sequences, which is a mainstream among new coronaviruses that are spreading in India. For the D614G, it is reported that it promotes the open conformation of RBD, and as a result, the infectivity of the new coronavirus is increased.
R408I, E471Q is said to be a mutation that is seen only in India.