A group from Department of Biology, University of York, York, UK, etc. has reported about Ralstonia solanacearum (the causative agent of bacterial wilt) inhibition by Pseudomonas strains and its underlying mechanisms of potential pathogen suppression.
https://onlinelibrary.wiley.com/doi/10.1002/mbo3.1283
It was found that Pseudomonas protegens CHA0 was the most inhibitory biocontrol strain against Ralstonia solanacearum.
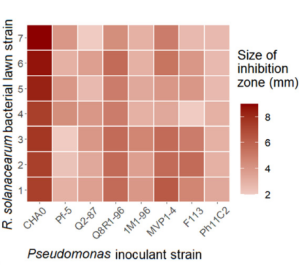
Secondary metabolite clusters were analyzed by using antiSMASH, and from 11 to 17 metabolic clusters were identified in each of the eight Pseudomonas genomes. Nonribosomal peptide synthetases (NRPS) were the most abundant secondary metabolite clusters. Similarly, DAPG metabolite (belonging to the T3PKS cluster), as well as the pyoverdine siderophore (NRPS cluster) metabolite clusters, were found in all strains. Overall, the highest number of clusters were detected in CHA0 and Pf-5 strains. These strains also harbored some unique metabolite clusters such as the T1PKS metabolic cluster, which encodes pyoluteorin antimicrobial, and the CDPS cluster, which encodes unknown metabolites. CHA0 and Pf-5 also had the greatest number of NRPS clusters and were the only two strains capable of producing the cyclic lipopeptides known as orfamides, and only orfamide A was production by CHA0 strain.
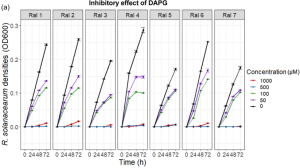
DAPG suppressed every R. solanacearum strain in a concentration-dependent manner, and all tested strains were unable to grow at the two highest concentrations (500 and 1000 μM).
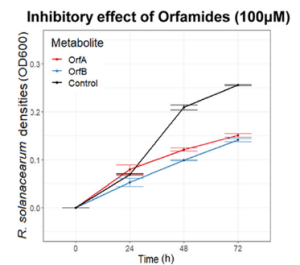
Orfamide variants “A” and “B” isolated from the Pseudomonas CHA0 strain were tested against R solanacearum strains (only #1 and #7 due to limited quantities of extracted compounds). Orfamide variants suppressed both Ralstonia strains as well.
It is quite likely that secondary metabolites from Pseudomonas strains such as DAPG and Orfamides could be pathogen-suppressing materials against R. solanacearum.
