A group from Lab Microbial Ecology of the Rhizosphere (LEMiRE), CEA, CNRS, BIAM, Aix Marseille Univ, Saint-Paul-Lez-Durance, France, etc. has reported about the complexity of the multiple molecular dialogues that take place underground between microorganisms and between plants and its rhizosphere microbiota.
https://sfamjournals.onlinelibrary.wiley.com/doi/10.1111/1751-7915.14023
In this report, it was investigated the ability of the plant root-associated Pseudomonas brassicacearum NFM421 known as phytobeneficial bacteria to compete with two other rhizobacteria, Kosakonia sacchari NO9, a root-associated diazotroph and Rhizobium alamii YAS34, known for its ability to produce exopolysaccharide. Especially, the expression of the well known phytobeneficial genes phlD (production of DAPG), hcnA (production of hydrogen cyanide) and acdS (ACC deaminase activity) was evaluated in a variety of situations. The intracellular status of iron in P. brassicacearum NFM421 was also evaluated by analysing the expression of iron-regulatory RNAs prrF in this strain, because when iron is scarce, bacteria produce siderophores that may act as antifungals by depriving fungi of iron. The antagonism of P. brassicacearum NFM421 wt and the knockout mutants ∆phlD and ∆gacA against the soil-borne plant pathogens Fusarium culmorum, Fusarium graminearum and Microdochium nivale was evaluated as well, to compare the antifungal activity of DAPG and HCN.
Pseudomonads promote plant growth and produce antimicrobial secondary metabolites including HCN and DAPG. The expression of phlD was one thousand times higher than that of hcnA, indicating that even if HCN is known to be effective against pathogenic fungi, the levels of its expression by P. brassicacearum NFM421 are too low to inhibit fungal growth. Therefore, the mode of biocontrol of plant pathogenic fungi used in this study, which seems plausible in P. brassicacearum NFM421, most likely involves DAPG.
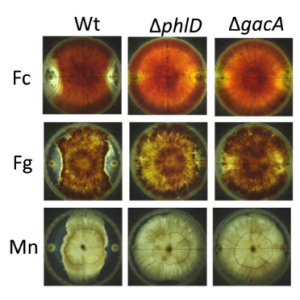 showing antifungal activity of P. brassicacearum NFM421wt, ∆phlD and ∆gacA towards Fusarium culmorum (Fc), Fusarium graminearum (Fg) or Microdochium nivale (Mn)
showing antifungal activity of P. brassicacearum NFM421wt, ∆phlD and ∆gacA towards Fusarium culmorum (Fc), Fusarium graminearum (Fg) or Microdochium nivale (Mn)
On in vitro conditions in CAA medium, the presence of the other two strains, N09 and YAS34, (separately or together) caused significant transcriptional changes in the expression of phlD, hcnA and acdS in P. brassicacearum NFM421 under iron-rich conditions. phlD was positively regulated by iron, an almost fourfold increase under iron-rich conditions, when the bacterium was grown alone. However, the presence of the competitors in CAA medium significantly decreased the phlD expression under iron-rich conditions (twofold), while no significant difference was observed under iron-depleted conditions. However, contrary to what was observed under in vitro conditions, phlD expression in P. brassicacearum NFM421 did not change during their interaction with B. napus root system in response to iron availability when grown alone.
As a conclusion, a scheme targeting the modulation of phl gene expression by other microorganisms and the plant was proposed, as well as the role of iron and its regulation by prrF RNAs. Anyway, the mechanisms by which one bacterial population interferes with the gene expression of another population are not yet fully understood and are future problems.
