A group from the National Institute of Child Health and Development demonstrated that deep learning can be combined with glycan profiling data from lectin microarrays to recognize cell differences with ultra-high accuracy.
https://www.sciencedirect.com/science/article/pii/S2352320420300742?via%3Dihub
The lectin microarray used was LecChip Ver1.0 of GlycoTechnica. The lectin microarray has 45 kinds of carefully selected natural lectins and has been widely used worldwide as a de facto standard for lectin arrays since its launch in 2007.
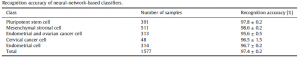
Deep Learning, as you know, is now used in various fields as one of methods of AI, and in this research, Google’s TensorFlow was used as the backend, and Keras was used as wrapper software. The layer configuration of this deep Leaning had an input layer of 45 (the same number of lectins) and an output layer of 5 (to discriminate five cells), and hidden layer from 1 to 5. There were five types of cells evaluated (Pluripotent stem cell, Mesenchymal stromal cell, Endometrial and ovarian cancer cell, Cervical cancer cell, Endometrial cell), and a total of 1,577 samples were used for the evaluation.
The results were astonishing as follows, and showed a high recognition accuracy of 97.4% overall.
Deep Learning software used in this paper is sold under the soft name “SA/DL Easy” from Mx. “SA/DL Easy” does not require any knowledge of programming such as Python, you can build neural networks and run deep learning just by clicking a mouse using a one-dimensional array dataset (such as glycan profiling data of lectin microarrays used in the above paper) as an input. “SA/DL Easy” is a quite user-friendly software to use Deep Learning. If you are interested in this software, please contact Mx.
