A group from Institute of Molecular Biotechnology of the Austrian Academy of Sciences, Vienna Austria, etc. has reported CLEC4G and CD209c lectins could block SARS-CoV-2 infections.
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC8420505/
Of 168 annotated carbohydrate recognition domains (CRDs) of mouse C‐type lectins, Galectins and Siglecs, 143 lectin‐CRDs as IgG2a‐Fc fusion proteins were expressed with human HEK293F cells. The resulting dimeric lectin‐Fc fusion proteins will be the first comprehensive library of mammalian CRDs. Through this study, the dimeric lectin-Fc fusion proteins were used as lectins.
The detected N‐glycan species from SARS-CoV-2 Spike ranged from poorly processed oligo‐mannose structures to highly processed multi‐antennary complex N‐glycans in a site‐dependent manner. The two glycosylation sites N331 and N343 located in the RBD carried more extended glycans, including sialylated and di‐fucosylated structures, when expressed as an independent construct as opposed to the full‐length Spike protein. It has to be noted that the N‐glycosylation of the RBD within full‐length trimeric Spike is different from N‐glycosylation of the RBD expressed as minimal ACE2 binding domain. This highlights the importance of using a full‐length trimeric Spike protein for its functional studies. This also suggests slight changes in glycosylation might explain differences in antiviral immunity and possibly severity of the disease.
Anyway, it was demonstrated that two lectins, CLEC4G and CD209c, were identified to strongly bind to Spike. CLEC4G and CD209c binding to Spike was visualized in real time and at single‐molecule resolution using atomic force microscopy, and the 3D modelling showed that both lectins can bind to a glycan within the RBD‐ACE2 interface and thus interferes with Spike binding to cell surfaces. Finally, it was shown that CLEC4G and CD209c significantly reduced SARS‐CoV‐2 infection.
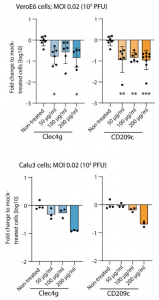 The Viral RNA was measured with qRT–PCR 15 h after infection of SARS‐CoV‐2. Data are presented as fold changes of viral loads over mock (only SARS‐CoV‐2 was added).
The Viral RNA was measured with qRT–PCR 15 h after infection of SARS‐CoV‐2. Data are presented as fold changes of viral loads over mock (only SARS‐CoV‐2 was added).
