A group from Department of Biotechnology, Delft University of Technology, Van der Maasweg 9, 2629 HZ Delft, The Netherlands, etc. has reported about changes of glycan composition in aerobic granular sludge by using lectin microarray and GC-MS etc.
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC10788855/
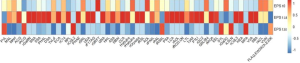
Aerobic granular sludge is a community of microbial organisms that remove carbon, nitrogen, phosphorus and other pollutants in a single sludge system used for wastewater treatments. In this research, the dynamic changes of the glycan profile of a few extracellular polymeric substance (EPS) samples extracted from aerobic granular sludge were monitored by GC-MS and lectin microarray.
EPSs were collected at different stages during its adaptation to the seawater condition on the initial day, 18th, and 30th days after the start of the reactor (denoted as EPSt0, EPSt18, and EPSt30 hereafter).
The application of the lectin microarray confirmed the presence of glycoproteins and effectively monitored its alteration along the adaptation to the seawater condition. Additionally, the result of lectin microarray is in line with the results of other analyses performed by GC-MS etc. This suggests that the lectin microarray is a successful platform for high-throughput glycan profiling of glycoproteins in microbial aggregates such as granular sludge.
Interestingly, there were more glycoproteins in EPSt18 than EPSt0 and EPSt30, and the glycosylation pattern of EPSt18 is significantly diverse. This indicates that, in response to the environmental change, i.e., exposure to the increased salt condition, one of the adaptation strategies of the microorganisms can be altering the glycosylation of proteins in quantity and diversity.
p.s., Her collaborator, Dr. Tateno in AIST, is also our collaborator regarding a glycan profiling system using lectin microarrays and an evanescent-filed fluorescence excitation scanner, GlycoStation.