SARS-CoV-2 Omicron showed higher infectivity in human nasal epithelial cells, but the reason is not only due to strong binding to hACE2
A group from Department of Infectious Disease, Imperial College London, UK, etc. has reported about SARS-CoV-2 Omicron’s higher infectivity in human nasal epithelial cells (hNECs) and its possible reasons.
https://www.biorxiv.org/content/10.1101/2021.12.31.474653v1
SARS-CoV-2 Omicron showed a large early replication advantage in human nasal epithelial cells (hNECs), yielding viral titres ~100-fold higher than Delta by 24 hours post-infection. At 48 and 72 hours post-infection, viral titres were lower compared to Delta and at 72 hours the RNA collected from Omicron infected wells was fewer. In Vero-AT cells, replication of the two variants was equal. In Calu-3 cells, the viral yields of Omicron were lower than for Delta across all time points.
Previous SARS-CoV-2 variants, such as Delta, only enter cells efficiently by binding ACE2 and activating fusion via cell-surface protease TMPRSS2. Omicron, conversely, is able to enter cells in both a TMPRSS2-dependent and –independent manner, having evolved the ability to avoid endosomal restriction. This allows Omicron to infect any ACE2-expressing cell in the airway instead of relying solely on double ACE+ TMPRSS2+ cells.
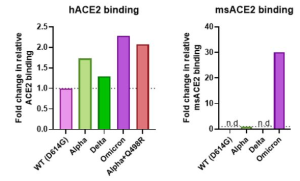
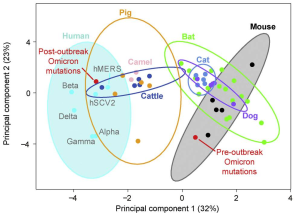
It was also found omicron Spike bound better to mouse ACE2 than any previous variant, as several others have reported. This suggests a possibility that omicron jumped from humans to mice, rapidly accumulated mutations conducive to infecting that host, then jumped back into humans.