Decay of Fc-dependent antibody functions in COVID-19 mild convalescent patients
A group from University of Melbourne, Australia, etc. has reported on decay of Fc-dependent antibody functions in COVID-19 mild convalescent patients.
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC8106889/
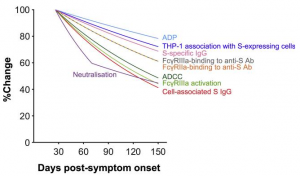
The FcγR-binding, antibody-dependent cellular cytotoxicity (ADCC), and antibody-dependent phagocytosis (ADP) activities of SARS-CoV-2 Spike-specific antibodies decay during convalescence from COVID-19. The first sample was collected at a median of 41 days post-symptom onset, and the last sample was collected at a median of 123 days post-symptom onset. The decline of plasma ADCC and ADP activity correlated with the decay of SARS-CoV-2 Spike-specific IgG and FcγR-binding antibodies (see figure below).
Importantly, Fc effector functions were readily detectable above uninfected controls in 94% of subjects for all assays at the last time point sampled, in contrast with neutralization activity, which remained detectable above background for only 70% of subjects. Overall, it seems that mild to moderate COVID-19 generates robust FcγR-binding, ADCC, and ADP antibody functions that decay at a slower rate than plasma neutralization activity.